Electron Transport Chains within the Nitrogen Cycle and the Carbon Cycle
Electron Transport Chains within the Nitrogen Cycle and the Carbon Cycle
Description. The following six figures represent electron transport chains functioning within the processes of nitrification and denitrification within the nitrogen cycle, and methanogenesis within the carbon cycle. Each figure is included with and without a legend. Figure 1 represents oxidation of ammonia to nitrite, the first phase of nitrification. Figure 2 represents oxidation of nitrite to nitrate, the second phase of nitrification. Figure 3 represents reduction of nitrate to molecular nitrogen through denitrification. Figure 4 represents oxidation of ammonia to nitrous oxide by ammonia-oxidizing bacteria. Figure 5 represents reduction of nitrate to nitrous oxide by incomplete denitrification. Figure 6 represents reduction of carbon dioxide by hydrogen gas to produce methane.
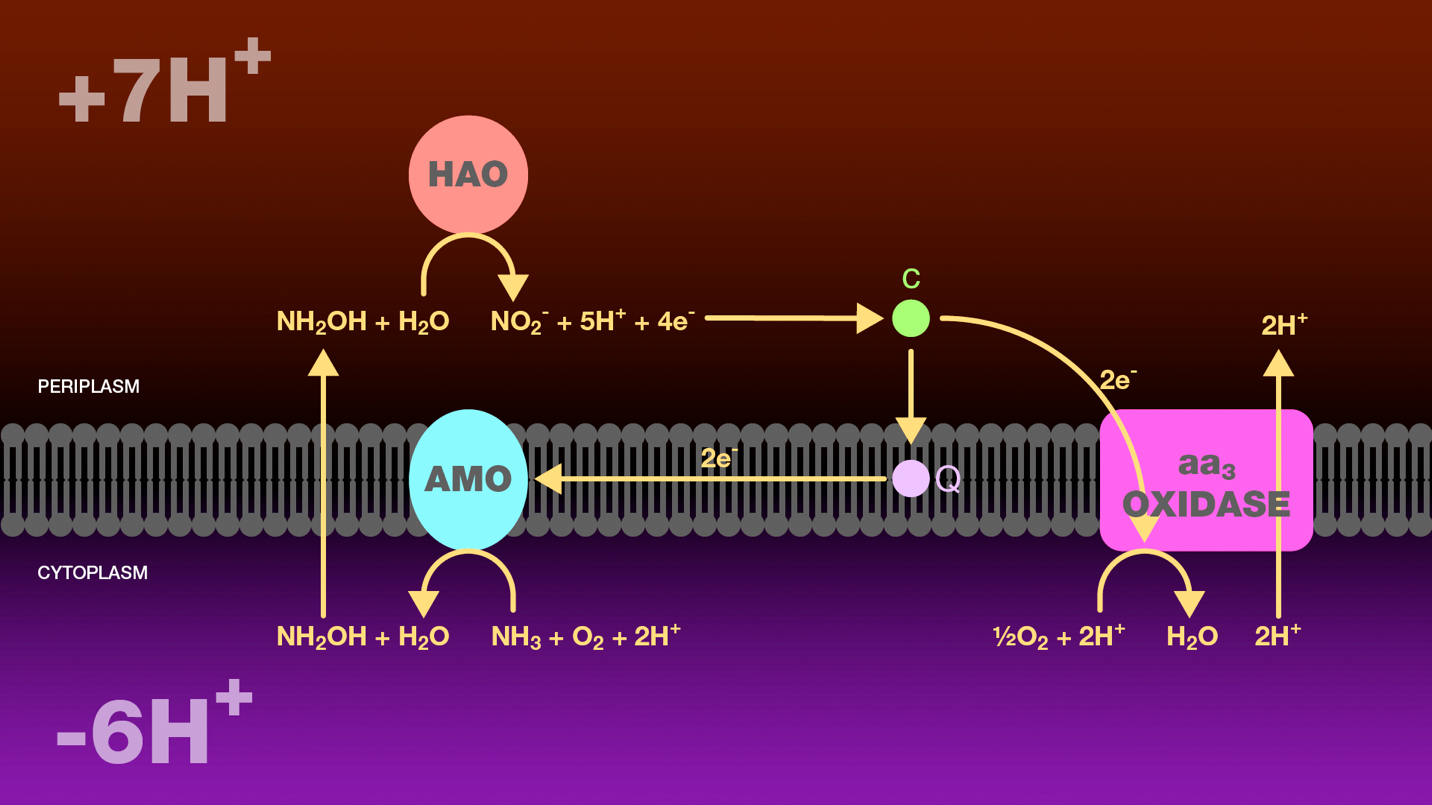
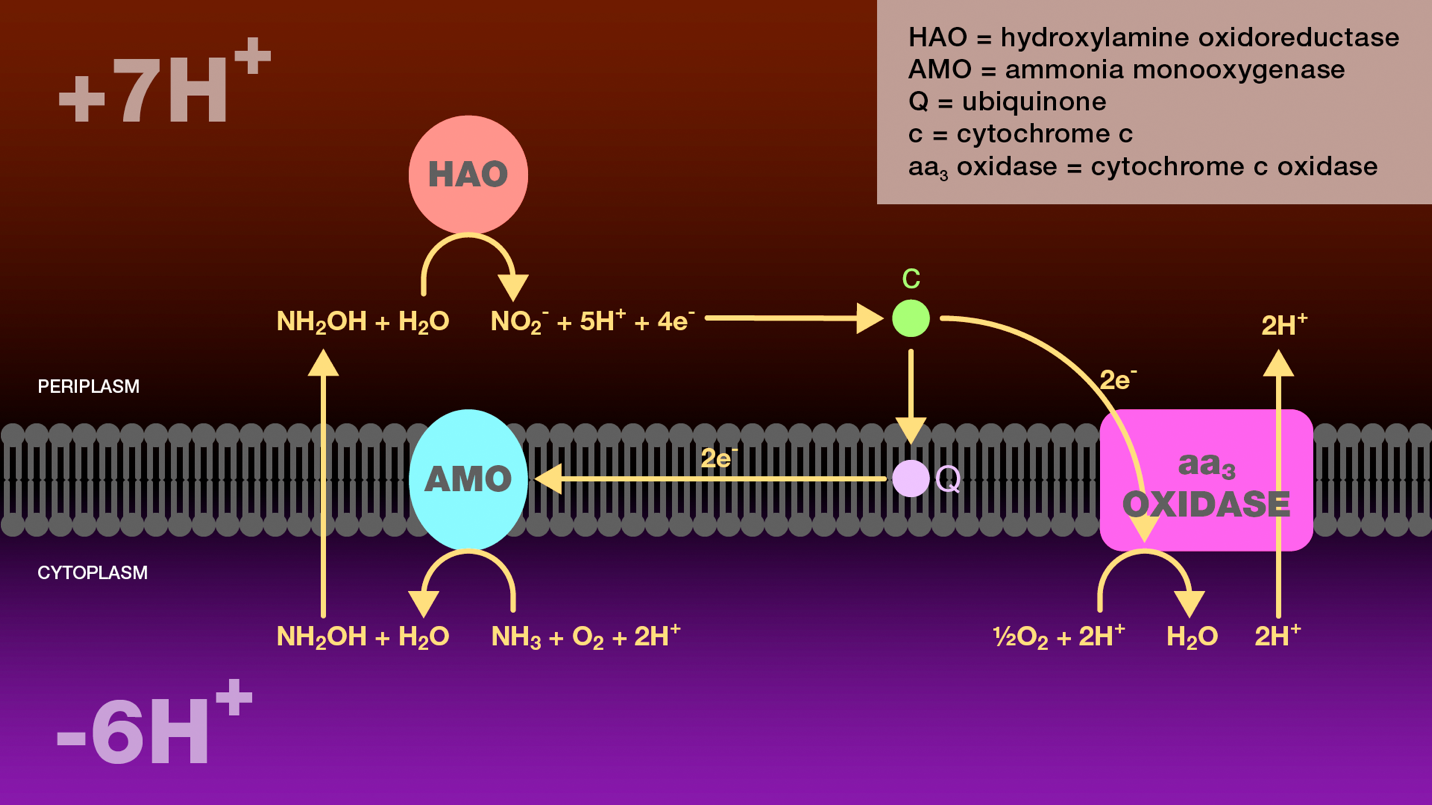
Figure 1. Oxidation of ammonia to nitrite by ammonia-oxidizing bacteria. Within the cytoplasm, ammonia monooxygenase catalyzes the oxidation of ammonia to hydroxylamine. Hydroxylamine diffuses across the inner bacterial membrane into the periplasm where it is further oxidized to nitrite by hydroxylamine oxidoreductase. Four electrons are released through this oxidation. Two electrons are transferred through cytochrome c and ubiquinone to ammonia monooxygenase to reduce molecular oxygen to water. The additional two electrons are transferred through cytochrome c to cytochrome c oxidase. Cytochrome c oxidase reduces molecular oxygen to water and transports two protons from the cytoplasm to the periplasm. The proton gradient created as a result of this electron transport scheme is the sole source of energy for these bacteria.
HAO = hydroxylamine oxidoreductase
AMO = ammonia monooxygenase
Q = ubiquinone
c = cytochrome c
aa3 oxidase = cytochrome c oxidase
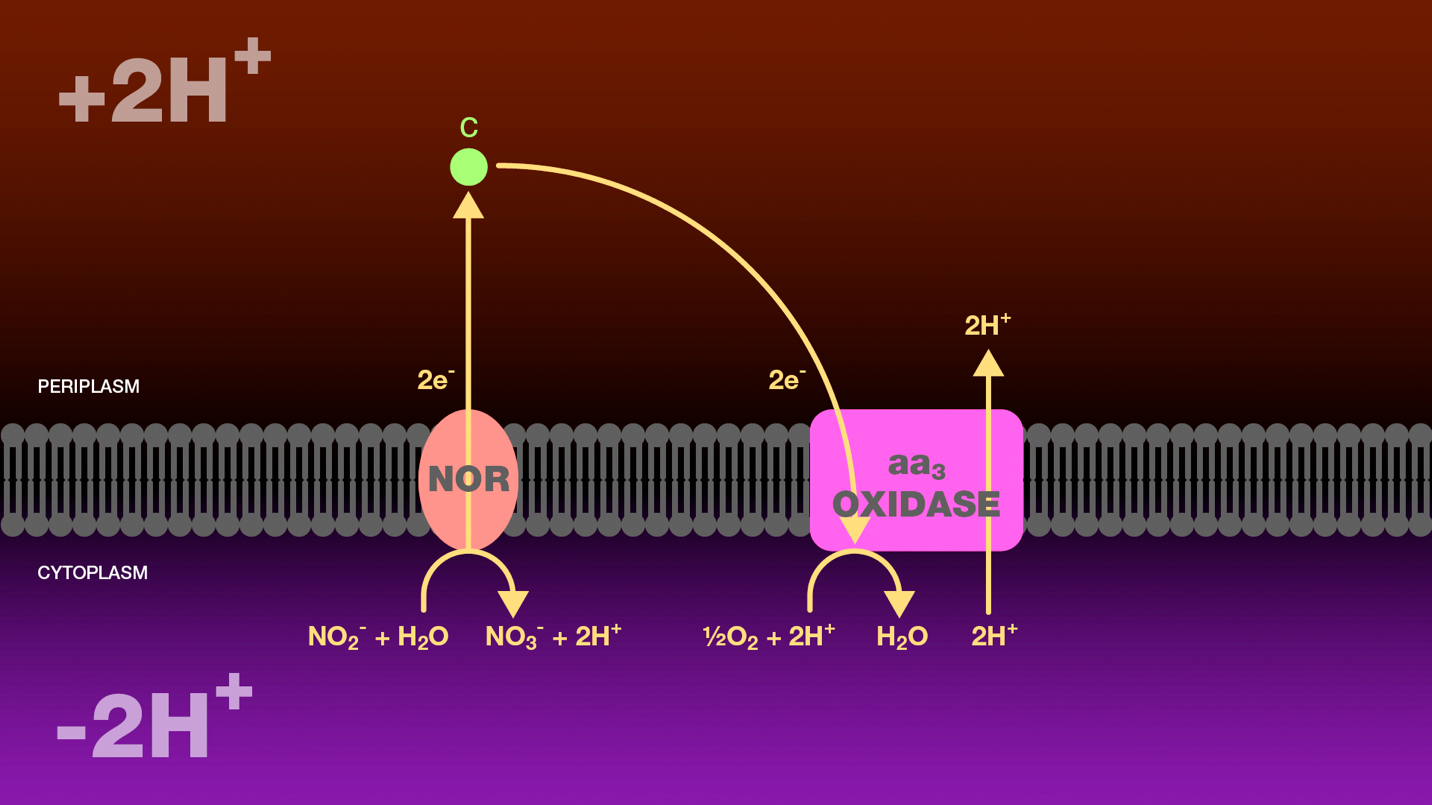
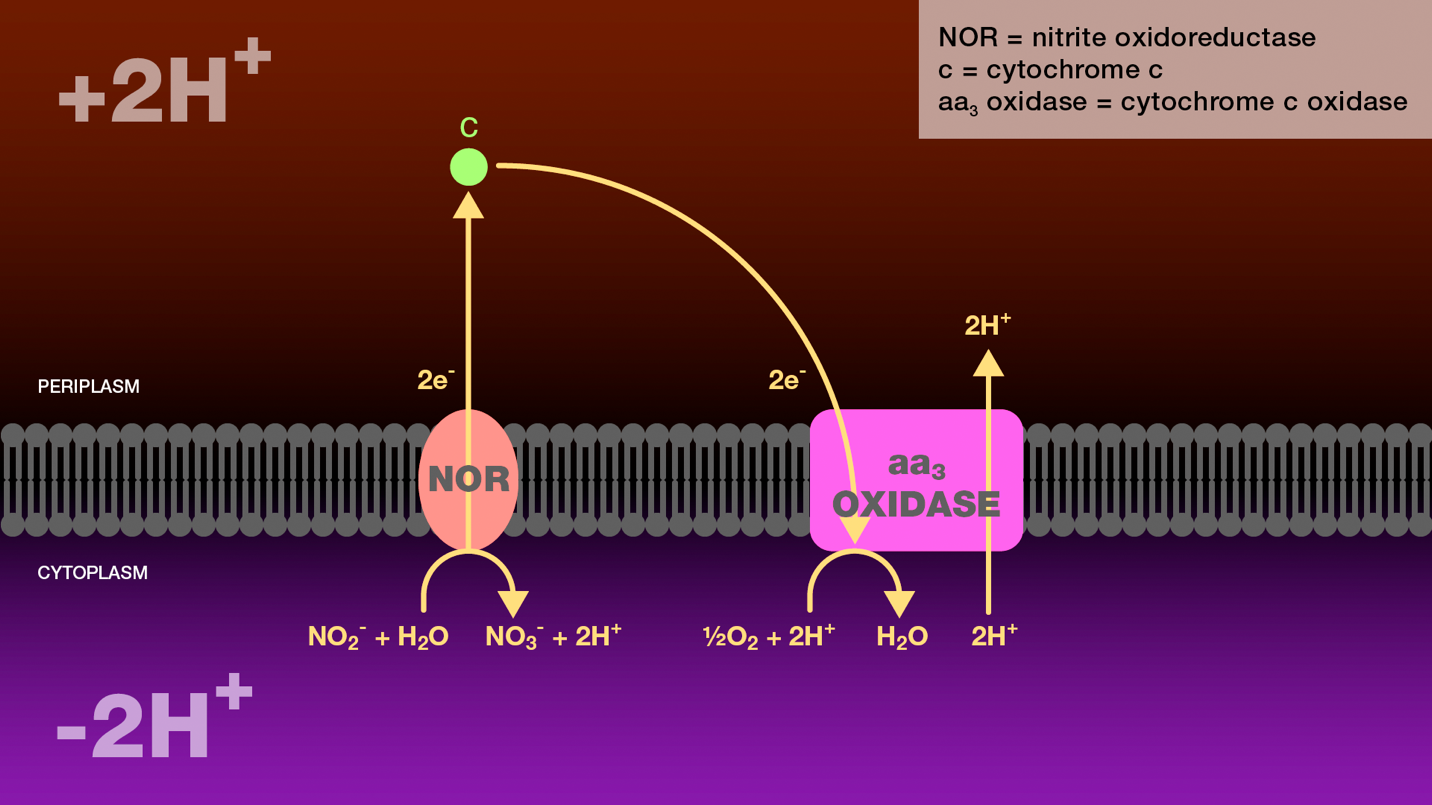
Figure 2. Oxidation of nitrite to nitrate by nitrite-oxidizing bacteria. Within the cytoplasm, nitrite oxidoreductase catalyzes the oxidation of nitrite to nitrate. Two electrons released during this oxidation are transferred through cytochrome c to cytochrome c oxidase. Cytochrome c oxidase reduces molecular oxygen to water and transports two protons from the cytoplasm to the periplasm. The proton gradient created as a result of this electron transport scheme is the sole source of energy for these bacteria.
NOR = nitrite oxidoreductase
c = cytochrome c
aa3 oxidase = cytochrome c oxidase
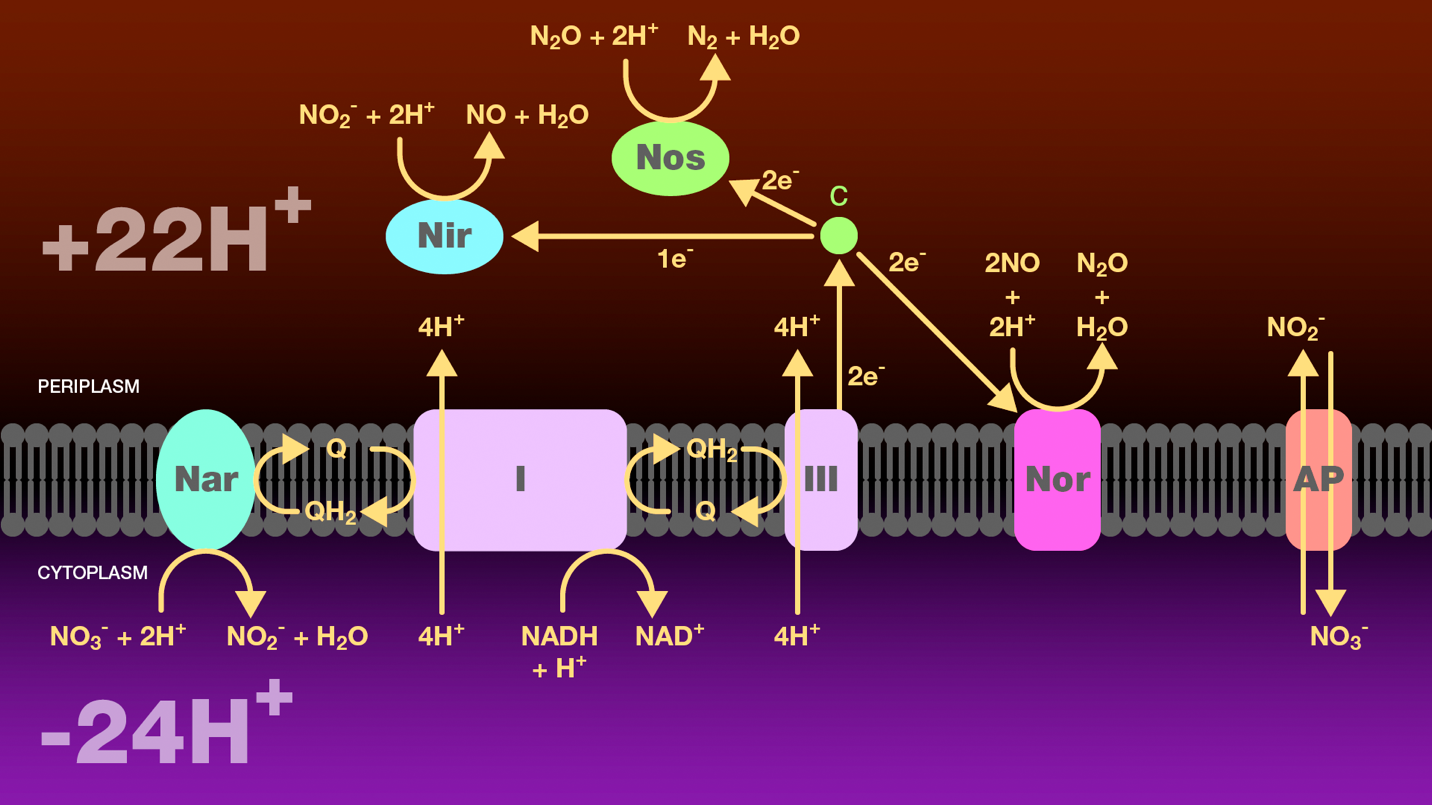
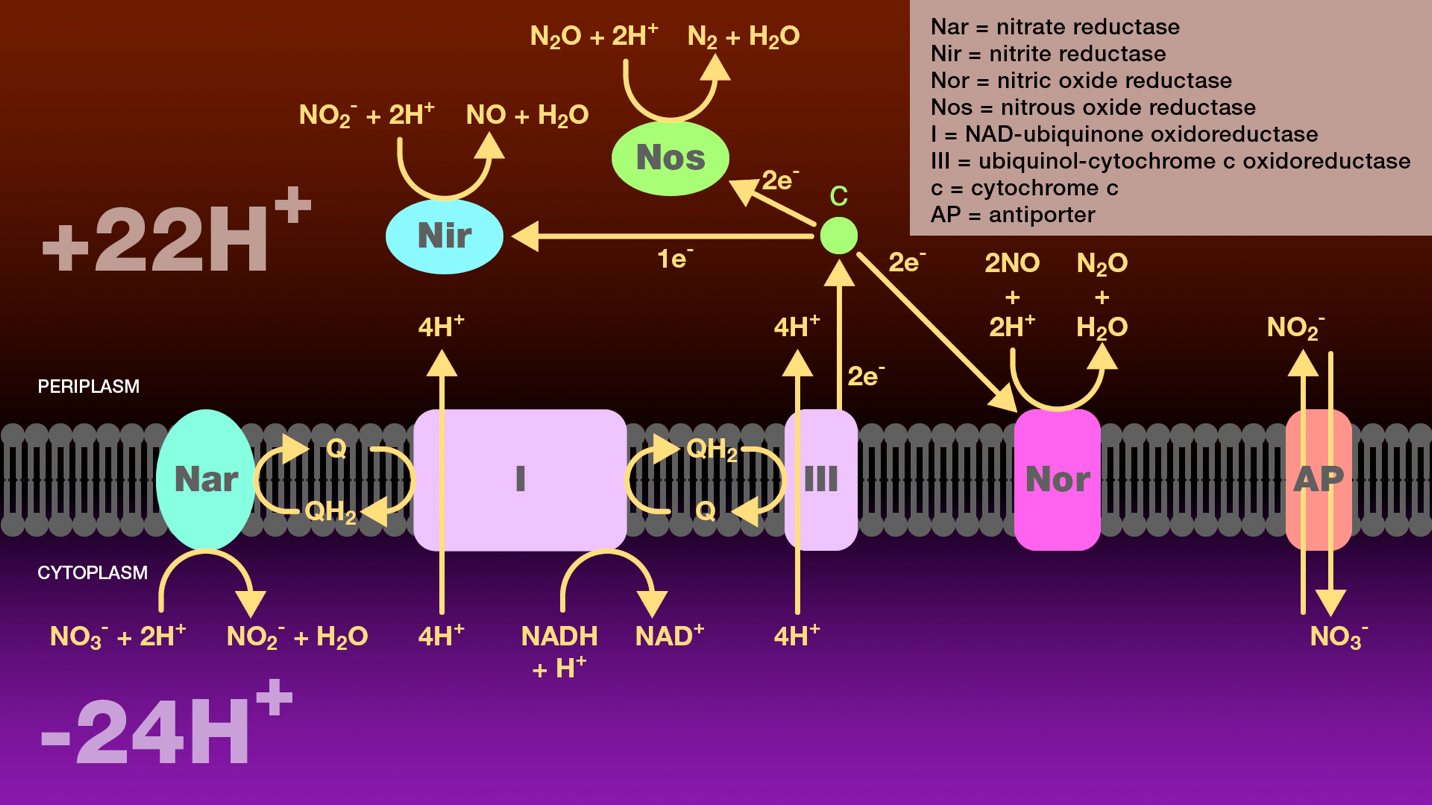
Figure 3. Reduction of nitrate to molecular nitrogen by denitrifying bacteria. A hydride ion from NADH and one proton from the cytosol are transferred to NADH-ubiquinone oxidoreductase and then to ubiquinol. NADH-ubiquinone oxidoreductase transfers four protons from the cytoplasm to the periplasm. Ubiquinol transfers two electrons either to nitrate reductase or to ubiquinol-cytochrome c oxidoreductase. Nitrate reductase catalyzes the reduction of nitrate to nitrite which is then transported to the periplasm through an antiporter. Ubiquinol-cytochrome c oxidoreductase transfers four protons from the cytoplasm to the periplasm and reduces cytochrome c. Cytochrome c is a branch point. It transfers one electron to nitrite reductase to catalyze the reduction of nitrate to nitric oxide, two electrons to nitric oxide reductase to catalyze the reduction of nitric oxide to nitrous oxide, and two electrons to nitrous oxide reductase to catalyze the reduction of nitrous oxide to molecular nitrogen. The proton gradient created as a result of this electron transport scheme provides a source of energy for these bacteria under anaerobic conditions.
Nar = nitrate reductase
Nir = nitrite reductase
Nor = nitric oxide reductase
Nos = nitrous oxide reductase
I = NAD-ubiquinone oxidoreductase
III = ubiquinol-cytochrome c oxidoreductase
c = cytochrome c
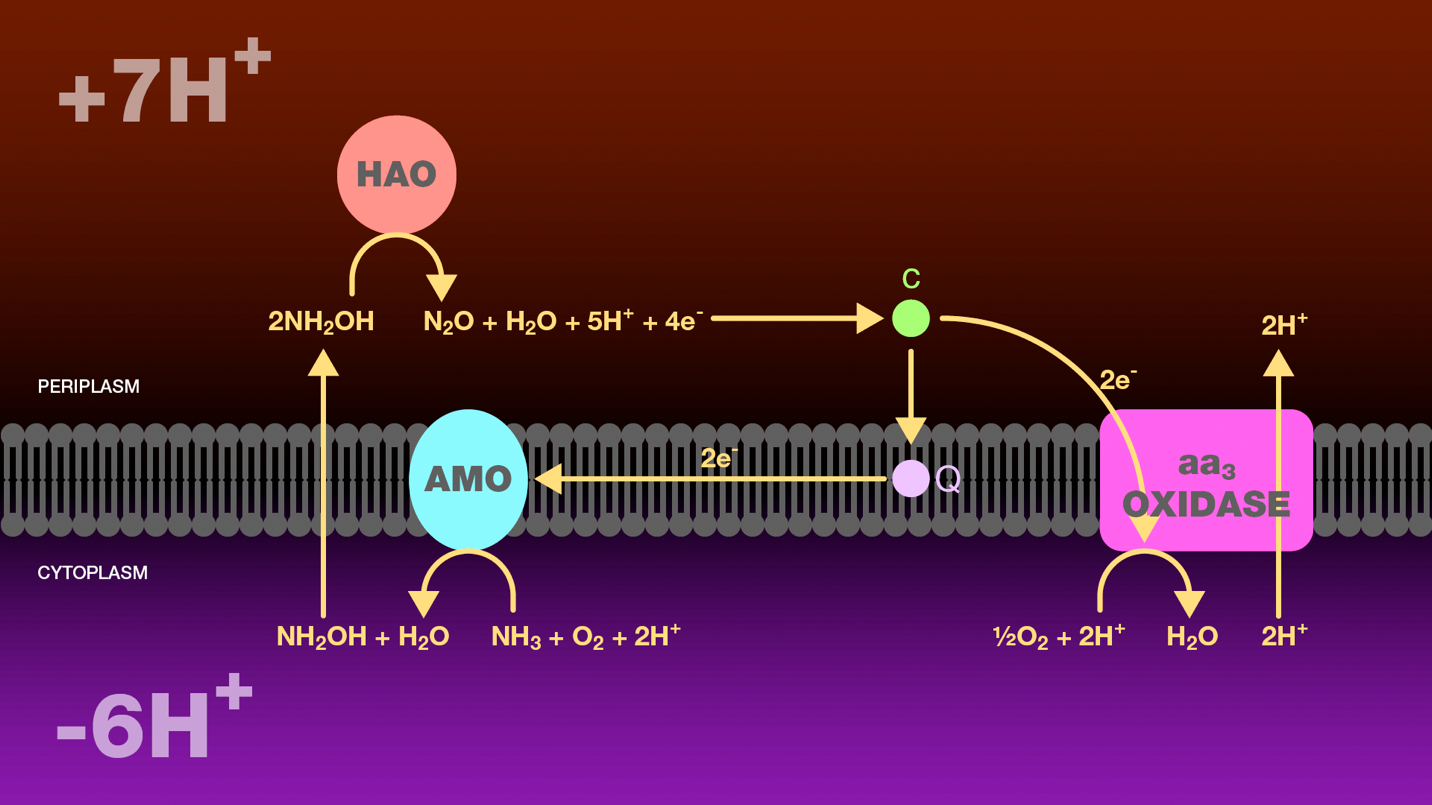
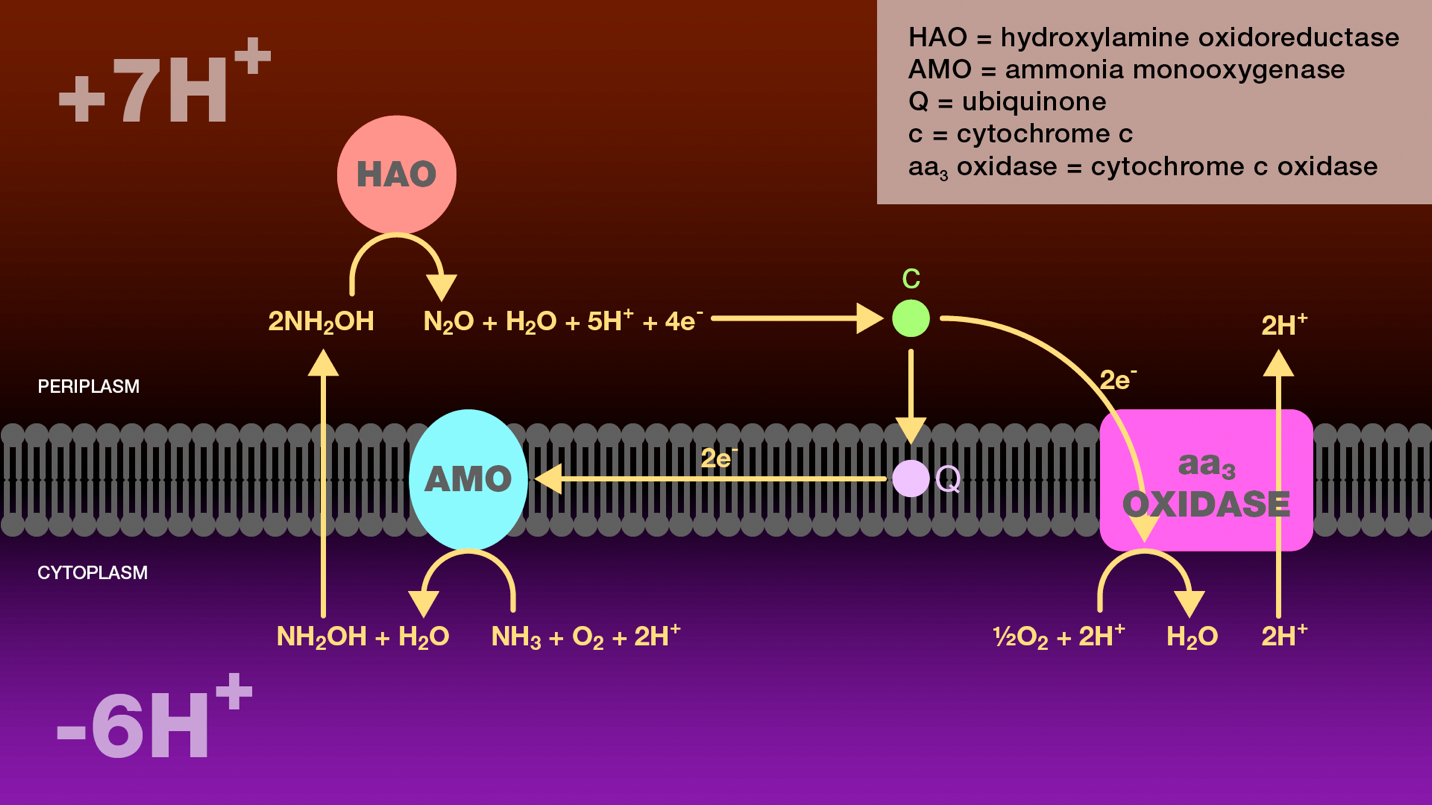
Figure 4. Oxidation of ammonia to nitrous oxide by ammonia-oxidizing bacteria. Within the cytoplasm, ammonia monooxygenase catalyzes the oxidation of ammonia to hydroxylamine. Hydroxylamine diffuses across the inner bacterial membrane into the periplasm where it is further oxidized to nitrous oxide by hydroxylamine oxidoreductase. Four electrons are released through this oxidation. Two electrons are transferred through cytochrome c and ubiquinone to ammonia monooxygenase to reduce molecular oxygen to water. The additional two electrons are transferred through cytochrome c552 to cytochrome c oxidase. Cytochrome c oxidase reduces molecular oxygen to water and transports two protons from the cytoplasm to the periplasm. The proton gradient created as a result of this electron transport scheme is the sole source of energy for these bacteria.
AMO = ammonia monooxygenase
HAO = hydroxylamine oxidoreductase
Q = ubiquinone
c554 = cytochrome c554
c552 = cytochrome c552
aa3 oxidase = cytochrome c oxidase
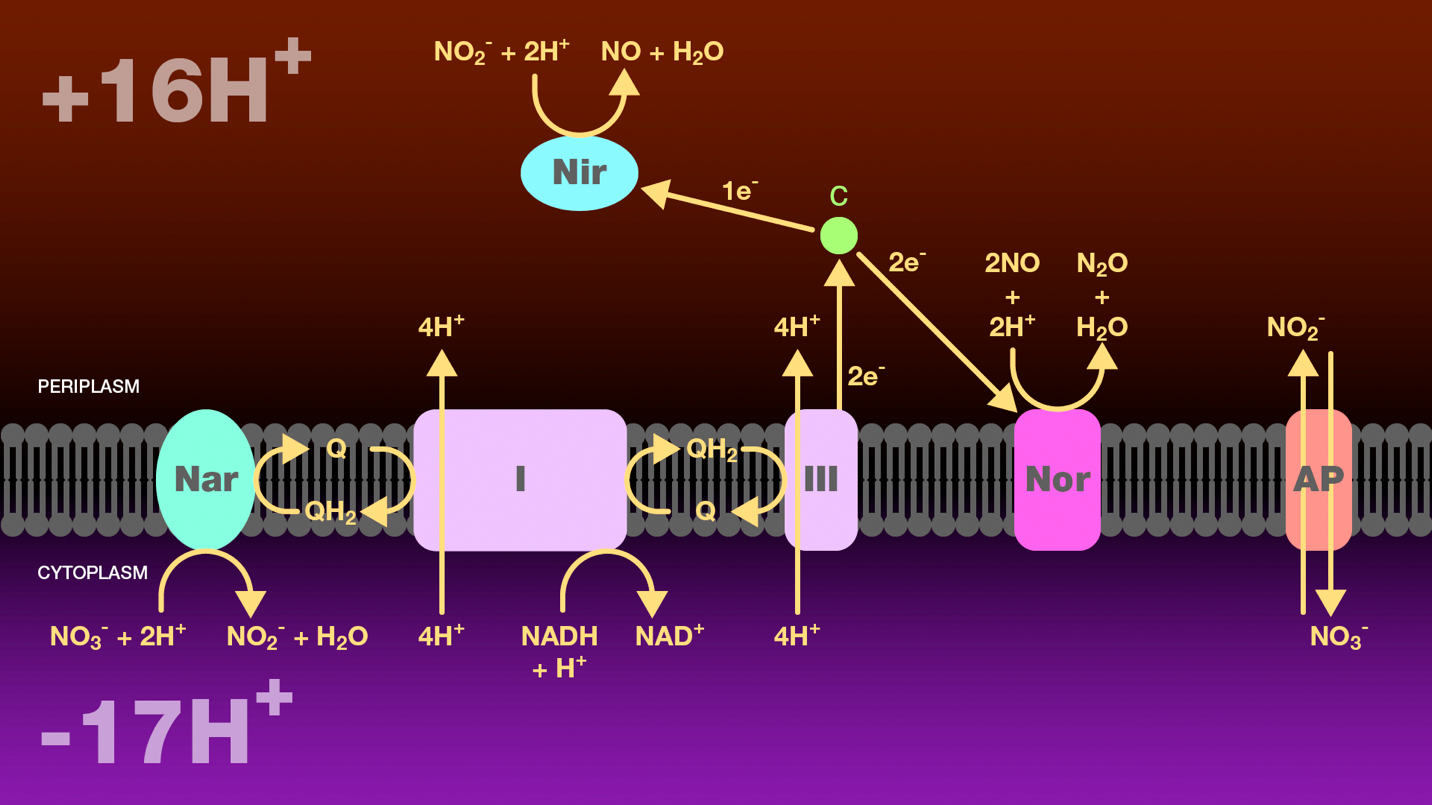
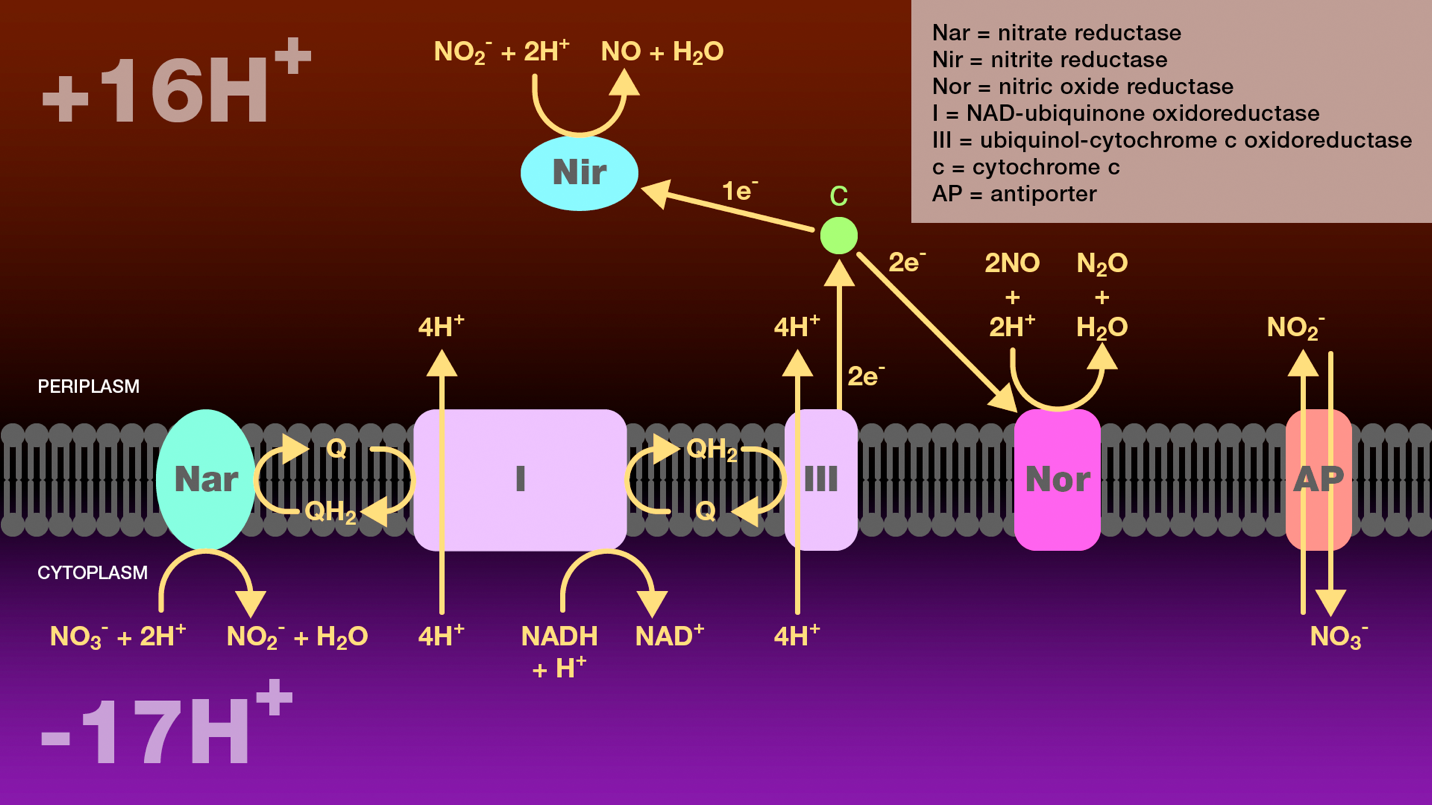
Figure 5. Reduction of nitrate to nitrous oxide by incomplete denitrification. A hydride ion from NADH and one proton from the cytosol are transferred to NADH-ubiquinone oxidoreductase and then to ubiquinol. NADH-ubiquinone oxidoreductase transfers four protons from the cytoplasm to the periplasm. Ubiquinol transfers two electrons either to nitrate reductase or to ubiquinol-cytochrome c oxidoreductase. Nitrate reductase catalyzes the reduction of nitrate to nitrite which is then transported to the periplasm through an antiporter. Ubiquinol-cytochrome c oxidoreductase transfers four protons from the cytoplasm to the periplasm and reduces cytochrome c. Cytochrome c is a branch point. It transfers one electron to nitrite reductase to catalyze the reduction of nitrate to nitric oxide, and two electrons to nitric oxide reductase to catalyze the reduction of nitric oxide to nitrous oxide. The proton gradient created as a result of this electron transport scheme provides a source of energy for these bacteria under anaerobic conditions.
Nar = nitrate reductase
Nir = nitrite reductase
Nor = nitric oxide reductase
I = NAD-ubiquinone oxidoreductase
III = ubiquinol-cytochrome c oxidoreductase
c = cytochrome c
AP = antiporter
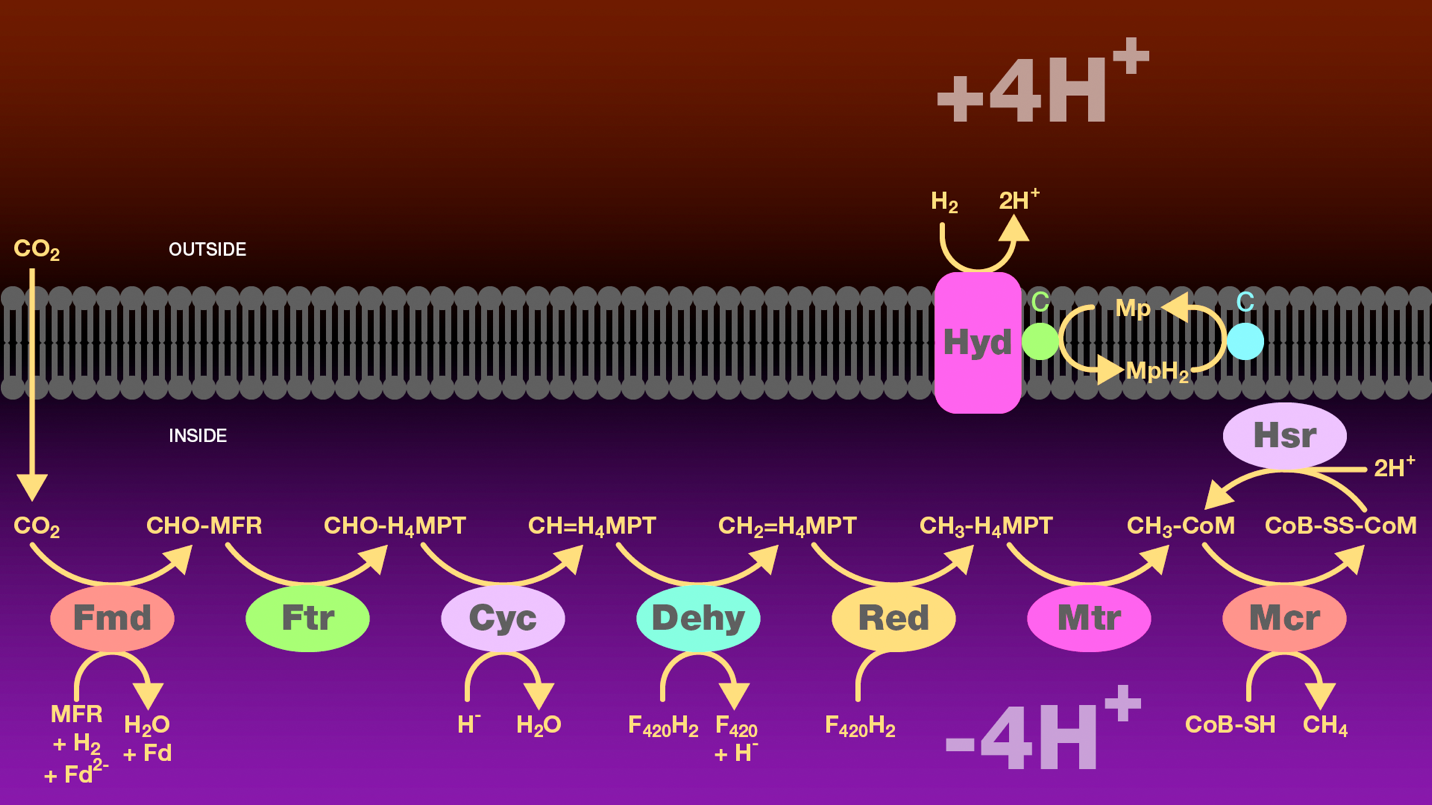
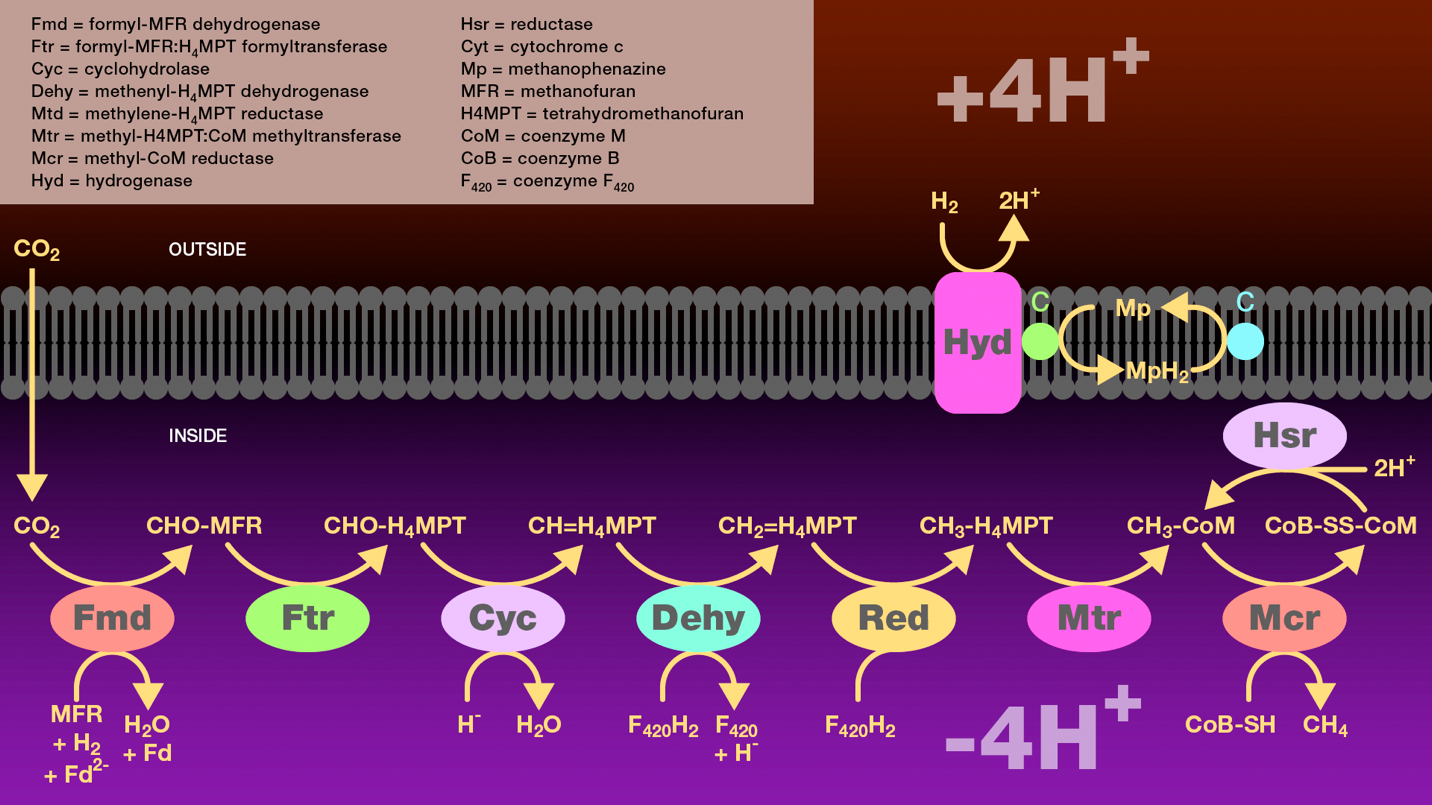
Figure 6. Hydrogenotrophic methanogenesis pathway. Carbon dioxide diffuses across the archaeal membrane and is reduced to methyl-coenzyme M. This process has six steps. The first step involves reductive activation of carbon dioxide to produce formylmethanofuran catalyzed by formylmethanofuran dehydrogenase. The second step involves transfer of the formyl group from methanofuran to tetrahydromethanopterin catalyzed by formylmethanofuran:tetrahydromethanofuran formyltransferase. The third step involves conversion of formyl-tetrahydromethanopterin to methenyl-tetrahydromethanopterin catalyzed by a cyclohydrolase. The fourth step involves reduction of methenyl-tetrahydromethanopterin to methylene-tetrahydromethanopoterin catalyzed by methenyltetrahydromethanopterin dehydrogenase. The fifth step involves reduction of methylene-tetrahydromethanopterin to methyl-tetrahydromethanopterin catalyzed by methylenetetrahydromethanopterin reductase. The sixth step involves transfer of the methyl group from tetrahydromethanopterin to coenzyme M catalyzed by methyltetrahydromethanopterin:coenzyme M methyltransferase. The methyl group is then reduced to methane by coenzyme B catalyzed by methyl-coenzyme M reductase. Finally, electrons are required to regenerate the reduced disulfide. Molecular hydrogen is oxidized by a hydrogenase and two electros are passed through cytochrome c and methanophenazine to a cytosolic reductase. This electron transport scheme generates a proton gradient across the archaeal membrane which is a source of energy for these methanogens.
Fmd = formyl-MFR dehydrogenase
Ftr = formyl-MFR:H4MPT formyltransferase
Cyc = cyclohydrolase
Dehy = methenyl-H4MPT dehydrogenase
Mtd = methylene-H4MPT reductase
Mtr = methyl-H4MPT:CoM methyltransferase
Mcr = methyl-CoM reductase
Hyd = hydrogenase
Hsr = reductase
Cyt = cytochrome c
Mp = methanophenazine
MFR = methanofuran
H4MPT = tetrahydromethanofuran
CoM = coenzyme M
CoB = coenzyme B
F420 = coenzyme F420
